Beyond 2D: A Deep Dive into Molecular Fingerprints for Capturing Stereochemistry - From Morgan to MapChiral
Published:
Encoding stereochemistry in molecular representations is critical for building robust QSAR and QSPR models, especially when predicting biological activity or physicochemical properties of chiral compounds. In this post, we delve into why stereochemistry matters, how different molecular fingerprints handle it, and what our practical results reveal about their effectiveness in distinguishing between enantiomers such as R- and S-thalidomide.
What is QSAR, and why does stereochemistry matter?
QSAR/QSPR (Quantitative Structure-Activity/Property Relationship) modeling is a computational framework for predicting the biological effects or properties of molecules based on their chemical structure. By correlating molecular features (descriptors or fingerprints) with experimental outcomes, QSAR accelerates drug discovery and toxicological screening, reducing the need for costly experiments. According to OECD guidelines, reliable QSAR models are essential tools for regulatory and industrial applications.
But not all molecular features are created equal-stereochemistry, the 3D arrangement of atoms, can have a dramatic impact on biological activity. Different stereoisomers (such as enantiomers or diastereomers) often bind to biological targets with different affinities, leading to large variations in drug efficacy, safety, or even toxicity.
The Real-world impact of stereochemistry
A classic example is thalidomide: its R-enantiomer is a sedative, while the S-enantiomer caused devastating birth defects, leading to one of the biggest tragedies in pharmaceutical history. Similarly, limonene's R-enantiomer smells like orange, while the S-enantiomer has a lemon scent-subtle 3D differences, big real-world consequences.
A metaphor: imagine giving 30 students a key to their classroom. Fifteen keys are exact copies (R-enantiomers), and the other fifteen are mirror images (S-enantiomers). Only the correct copies open the classroom door; the mirror-image keys might fit somewhere else, possibly with unintended outcomes.
This perfectly illustrates why distinguishing between stereoisomers is crucial in cheminformatics and drug design.
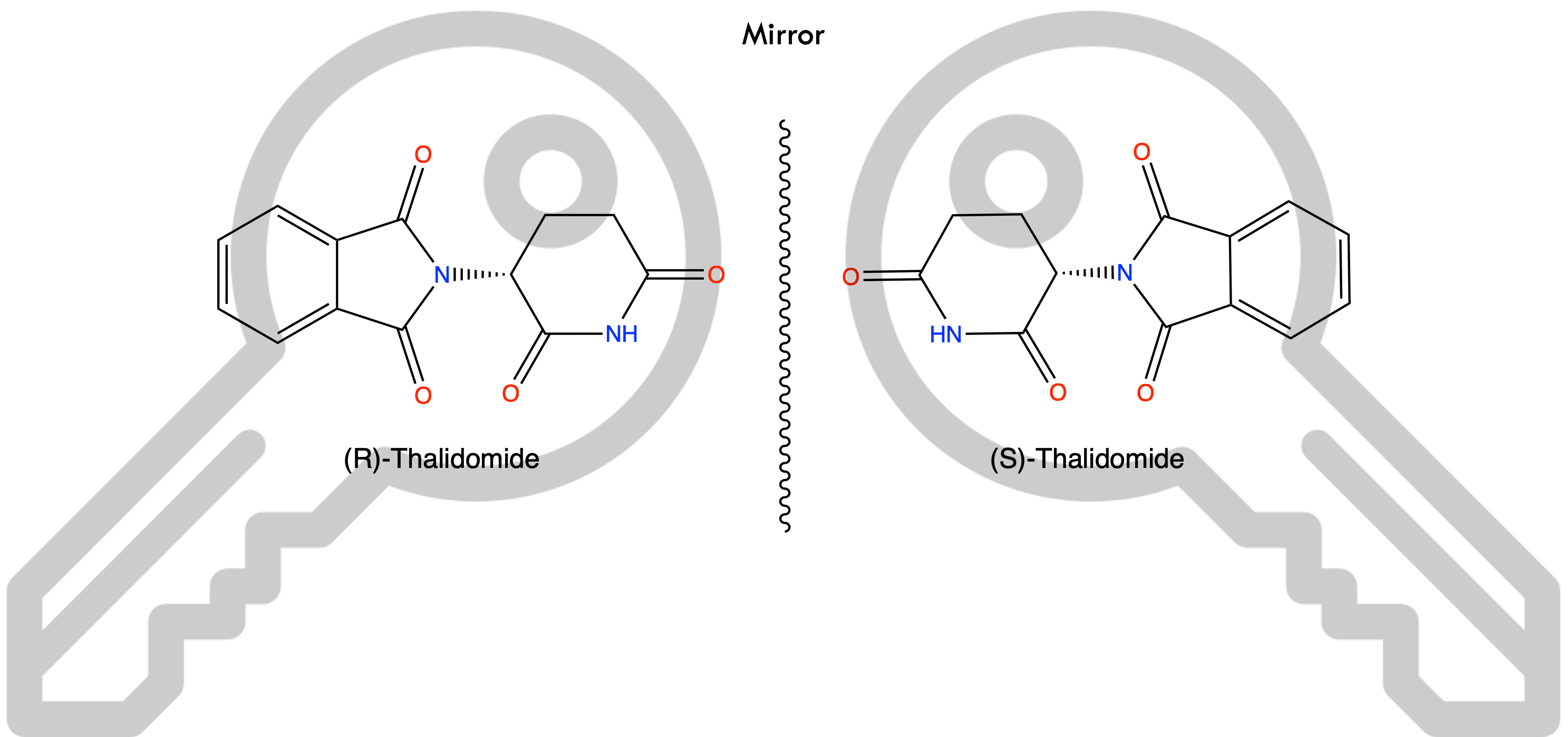
The Challenge: Representing stereochemistry for ML
Capturing stereochemistry in molecular descriptors is not trivial. Traditional QSAR descriptors are often 2D and may miss stereochemical differences entirely, leading to models that cannot distinguish between enantiomers. Fully 3D approaches (like CoMFA or CoMSIA) account for stereochemistry but require reliable conformer generation and alignment, which is computationally intensive and less suited to large, diverse datasets.
A practical and scalable solution is to use 2D fingerprints that encode stereochemistry. The sensitivity of different fingerprints to stereochemical differences varies, and choosing the right one is key for robust and interpretable modeling.
Practical comparison: How well do fingerprints capture stereochemistry?
mapchiral library, generating several types of fingerprints:- Morgan (ECFP) with chirality enabled (radius 1 and 2)
- RDKit fingerprint
- MACCS keys
- Topological Torsion (with
includeChirality=True) - MapChiral (MinHashed Atom-Pair Chiral)
We then calculated the similarity between the fingerprints of the two enantiomers at different radii for Morgan and MapChiral. (Note: Topological Torsion does not use a radius parameter.) If a fingerprint is sensitive to stereochemistry, the similarity between R- and S-thalidomide should be less than 1.0.
from rdkit import Chem
from rdkit.Chem import AllChem, MACCSkeys, DataStructs, rdFingerprintGenerator
from mapchiral.mapchiral import encode, jaccard_similarity
# Define SMILES for R- and S-thalidomide
smiles_R = "O=C(N1)CC[C@@H](N2C(C3=CC=CC=C3C2=O)=O)C1=O"
smiles_S = "O=C(N1)CC[C@H](N2C(C3=CC=CC=C3C2=O)=O)C1=O"
mol_R = Chem.MolFromSmiles(smiles_R)
mol_S = Chem.MolFromSmiles(smiles_S)
# Generate Morgan fingerprints with different radii
def get_morgan_fp(mol, r):
mfpgen = rdFingerprintGenerator.GetMorganGenerator(radius=r, fpSize=2048, includeChirality=True)
return mfpgen.GetFingerprint(mol)
# Generate MapChiral fingerprints with different radii
def get_mapchiral_fp(mol, r):
return encode(mol, max_radius=r, n_permutations=2048, mapping=False)
# Generate Topological Torsion fingerprint with chirality
def get_torsion_fp(mol):
ttgen = rdFingerprintGenerator.GetTopologicalTorsionGenerator(fpSize=2048, includeChirality=True)
return ttgen.GetFingerprint(mol)
# Other fingerprints
def get_other_fps(mol):
fps = {}
fps['RDKit'] = Chem.RDKFingerprint(mol)
fps['MACCS'] = MACCSkeys.GenMACCSKeys(mol)
return fps
# Similarity functions
def tanimoto(fp1, fp2):
return DataStructs.TanimotoSimilarity(fp1, fp2)
def dice(fp1, fp2):
return DataStructs.DiceSimilarity(fp1, fp2)
fps_R1 = get_morgan_fp(mol_R, 1)
fps_S1 = get_morgan_fp(mol_S, 1)
fps_R2 = get_morgan_fp(mol_R, 2)
fps_S2 = get_morgan_fp(mol_S, 2)
mapchiral_R1 = get_mapchiral_fp(mol_R, 1)
mapchiral_S1 = get_mapchiral_fp(mol_S, 1)
mapchiral_R2 = get_mapchiral_fp(mol_R, 2)
mapchiral_S2 = get_mapchiral_fp(mol_S, 2)
torsion_R = get_torsion_fp(mol_R)
torsion_S = get_torsion_fp(mol_S)
other_fps_R = get_other_fps(mol_R)
other_fps_S = get_other_fps(mol_S)
# Compute similarities
print("Similarity between R- and S-thalidomide:")
print(f"Morgan fingerprint (radius 1) similarity: {tanimoto(fps_R1, fps_S1):.3f}")
print(f"Morgan fingerprint (radius 2) similarity: {tanimoto(fps_R2, fps_S2):.3f}")
print(f"MapChiral fingerprint (radius 1) similarity: {jaccard_similarity(mapchiral_R1, mapchiral_S1):.3f}")
print(f"MapChiral fingerprint (radius 2) similarity: {jaccard_similarity(mapchiral_R2, mapchiral_S2):.3f}")
print(f"Topological Torsion fingerprint similarity: {dice(torsion_R, torsion_S):.3f}")
for key in ['RDKit', 'MACCS']:
sim = tanimoto(other_fps_R[key], other_fps_S[key])
print(f"{key} fingerprint similarity: {sim:.3f}")
Output:
Similarity between R- and S-thalidomide:
Morgan fingerprint (radius 1) similarity: 0.900
Morgan fingerprint (radius 2) similarity: 0.714
MapChiral fingerprint (radius 1) similarity: 0.745
MapChiral fingerprint (radius 2) similarity: 0.879
Topological Torsion fingerprint similarity: 0.635
RDKit fingerprint similarity: 1.000
MACCS fingerprint similarity: 1.000
What do these results mean? (Radius 1 vs. Radius 2)
- Morgan (chiral) fingerprint: At radius 1, the similarity is 0.900, indicating moderate sensitivity to stereochemistry. At radius 2, the similarity drops significantly to 0.714, showing substantially improved ability to distinguish enantiomers as the fingerprint encompasses more extended molecular environments.
- MapChiral fingerprint: At radius 1, the similarity is 0.745 demonstrating strong sensitivity to chirality at a local level. At radius 2, the similarity rises to 0.879, reflecting the broader atom-pair relationships being encoded.
- Topological Torsion fingerprint (with chirality): The similarity is 0.635, showing that enabling chirality allows this fingerprint to distinguish enantiomers, though the value is even lower than both Morgan and MapChiral at radius 1.
- RDKit and MACCS fingerprints: Both return perfect similarity (1.000), confirming their inability to distinguish between enantiomers.
Interpreting the results: How well do fingerprints capture stereochemistry?
| Fingerprint Type | Radius | Similarity Score | Stereochemistry Sensitivity? |
|---|---|---|---|
| Morgan (chiral) | 1 | 0.900 | Partial |
| Morgan (chiral) | 2 | 0.714 | High (captures extended chirality) |
| MapChiral (MinHashed) | 1 | 0.745 | High (captures global chirality) |
| MapChiral (MinHashed) | 2 | 0.879 | High (radius-dependent) |
| Topological Torsion | - | 0.635 | Yes (with chirality enabled) |
| RDKit | - | 1.000 | No |
| MACCS | - | 1.000 | No |
These results clearly show that Morgan (with chirality), Topological Torsion (with chirality), and MapChiral fingerprints can distinguish between R- and S-enantiomers of thalidomide, while classical RDKit and MACCS fingerprints cannot. The sensitivity of each fingerprint depends on its algorithm and parameters:
- MapChiral encodes atom-pair and chiral information globally, making it robust to the radius parameter and highly effective at distinguishing enantiomers, even when their overall structures are very similar.
- Morgan (chiral) fingerprints show improved discrimination as the radius increases, capturing more extended chiral environments.
- Topological Torsion (with chirality) also distinguishes enantiomers, sometimes even more stringently than Morgan in this example.
- RDKit and MACCS fingerprints return perfect similarity (1.000), confirming their inability to capture stereochemical differences.
Scientific takeaways and best practices
- Always use stereochemistry-sensitive fingerprints (such as Morgan with
includeChirality=True, Topological Torsion with chirality, or MapChiral) for ML/QSAR tasks involving chiral molecules. - Be aware of the limitations of classical fingerprints: RDKit and MACCS cannot distinguish between enantiomers and should not be used where stereochemistry matters.
- Adjust fingerprint parameters thoughtfully: For Morgan fingerprints, changing the radius can improve stereochemical sensitivity, but the optimal setting depends on the molecular context.
- MapChiral offers robust chiral discrimination: It encodes atom-pair relationships and chirality globally, making it especially powerful for large, diverse datasets with many chiral centers.
- You don’t always need 3D QSAR: With the right 2D chiral-sensitive fingerprints, you can efficiently incorporate stereochemical information, avoiding the computational burden and uncertainty of 3D conformer generation.
Final remarks
Incorporating stereochemistry into molecular representations is not just a technical nuance-it can be the difference between a safe, effective drug and a harmful one. As our results show, using advanced chiral-aware fingerprints like MapChiral, Topological Torsion (with chirality), or properly configured Morgan fingerprints is crucial for building predictive, interpretable models in cheminformatics and drug design. Whenever chirality matters, choose your molecular representation wisely and avoid the pitfalls of stereochemistry ignorance!. So, with easy and straightforward approaches to encoding chiral information, we can enhance the reliability of our models and ultimately improve drug discovery outcomes.
Leave a Comment
Have thoughts, questions, or experiences to share? Leave a comment below! Your feedback and discussion are welcome.
References:
- Todeschini, R., & Consonni, V. (2009). Molecular Descriptors for Chemoinformatics. Wiley-VCH.
- Riniker, S., & Landrum, G. A. (2013). Open-source platform for molecular informatics. J. Cheminformatics, 5, 26.
Share on
Bluesky Facebook LinkedIn X (formerly Twitter)Leave a Comment
Your email address will not be published. Required fields are marked *

Leave a Comment
Your email address will not be published. Required fields are marked *